The set of functions described below is designed to handle
the image-related data stored inside a SpatialExperiment's
imgData int_metadata field. These include:
getImg,addImg,rmvImgto retrieve/add/remove an image entry to/from theimgDataDataFrameimgSource,imgRasterto retrieve the path/URL andrasterobject, respectively, associated with an image or set of images
Usage
# S4 method for class 'SpatialExperiment'
getImg(x, sample_id = NULL, image_id = NULL)
# S4 method for class 'SpatialExperiment'
addImg(x, imageSource, scaleFactor, sample_id, image_id, load = TRUE)
# S4 method for class 'SpatialExperiment'
rmvImg(x, sample_id = NULL, image_id = NULL)
# S4 method for class 'SpatialExperiment'
imgSource(x, sample_id = NULL, image_id = NULL, path = FALSE)
# S4 method for class 'SpatialExperiment'
imgRaster(x, sample_id = NULL, image_id = NULL)
# S4 method for class 'SpatialExperiment'
rotateImg(x, sample_id = NULL, image_id = NULL, degrees = 90)
# S4 method for class 'SpatialExperiment'
mirrorImg(x, sample_id = NULL, image_id = NULL, axis = c("h", "v"))Arguments
- x
- sample_id
character string,
TRUEorNULLspecifying sample/image identifier(s); here,TRUEis equivalent to all samples/images andNULLspecifies the first available entry (see details)- image_id
see
sample_id- imageSource
a character string specifying an image file name (.png, .jpg or .tif) or URL to source the image from
- scaleFactor
single numeric scale factor used to rescale spatial coordinates according to the image's resolution
- load
logical; should the image(s) be loaded into memory as a
rasterobject? ifFALSE, will store the path/URL instead- path
logical; for
RemoteSpatialImages, TRUE returns the path to the image's cached file, and FALSE its URL. ForStored/LoadedSpatialImages, a path/NA is returned, irrespective ofpath.- degrees
single numeric in +/-[0,90,...,360] specifying how many degrees to rotate. A negative/positive value corresponds to counter-/clockwise rotation
- axis
character string specifying whether to mirror horizontally (
"h") or vertically ("v")
Value
getImg() returns a single or list of SpatialImage(s).
add/rmvImg() return a SpatialExperiment
with modified imgData; specifically, they create/remove
an image entry (row) in the imgData DataFrame.
imgRaster/Source() access relevant data in the
SpatialImage(s) stored inside the imgData's data
field. Depending on whether or not multiple entries are accessed,
a character string or vector is returned by imgSource(), and a
single or list of raster object(s) is returned by imgRaster().
rotate/mirrorImg() return a LoadedSpatialImage
with modified a raster matrix.
Examples
example(read10xVisium)
#>
#> rd10xV> dir <- system.file(
#> rd10xV+ file.path("extdata", "10xVisium"),
#> rd10xV+ package = "SpatialExperiment")
#>
#> rd10xV> sample_ids <- c("section1", "section2")
#>
#> rd10xV> samples <- file.path(dir, sample_ids, "outs")
#>
#> rd10xV> list.files(samples[1])
#> [1] "raw_feature_bc_matrix" "spatial"
#>
#> rd10xV> list.files(file.path(samples[1], "spatial"))
#> [1] "scalefactors_json.json" "tissue_lowres_image.png"
#> [3] "tissue_positions_list.csv"
#>
#> rd10xV> file.path(samples[1], "raw_feature_bc_matrix")
#> [1] "/tmp/RtmppdZi9A/temp_libpathcf263c794d1/SpatialExperiment/extdata/10xVisium/section1/outs/raw_feature_bc_matrix"
#>
#> rd10xV> (spe <- read10xVisium(samples, sample_ids,
#> rd10xV+ type = "sparse", data = "raw",
#> rd10xV+ images = "lowres", load = FALSE))
#> class: SpatialExperiment
#> dim: 50 99
#> metadata(0):
#> assays(1): counts
#> rownames(50): ENSMUSG00000051951 ENSMUSG00000089699 ...
#> ENSMUSG00000005886 ENSMUSG00000101476
#> rowData names(1): symbol
#> colnames(99): AAACAACGAATAGTTC-1 AAACAAGTATCTCCCA-1 ...
#> AAAGTCGACCCTCAGT-1 AAAGTGCCATCAATTA-1
#> colData names(4): in_tissue array_row array_col sample_id
#> reducedDimNames(0):
#> mainExpName: NULL
#> altExpNames(0):
#> spatialCoords names(2) : pxl_col_in_fullres pxl_row_in_fullres
#> imgData names(4): sample_id image_id data scaleFactor
#>
#> rd10xV> # base directory 'outs/' from Space Ranger can also be omitted
#> rd10xV> samples2 <- file.path(dir, sample_ids)
#>
#> rd10xV> (spe2 <- read10xVisium(samples2, sample_ids,
#> rd10xV+ type = "sparse", data = "raw",
#> rd10xV+ images = "lowres", load = FALSE))
#> class: SpatialExperiment
#> dim: 50 99
#> metadata(0):
#> assays(1): counts
#> rownames(50): ENSMUSG00000051951 ENSMUSG00000089699 ...
#> ENSMUSG00000005886 ENSMUSG00000101476
#> rowData names(1): symbol
#> colnames(99): AAACAACGAATAGTTC-1 AAACAAGTATCTCCCA-1 ...
#> AAAGTCGACCCTCAGT-1 AAAGTGCCATCAATTA-1
#> colData names(4): in_tissue array_row array_col sample_id
#> reducedDimNames(0):
#> mainExpName: NULL
#> altExpNames(0):
#> spatialCoords names(2) : pxl_col_in_fullres pxl_row_in_fullres
#> imgData names(4): sample_id image_id data scaleFactor
#>
#> rd10xV> # tabulate number of spots mapped to tissue
#> rd10xV> cd <- colData(spe)
#>
#> rd10xV> table(
#> rd10xV+ in_tissue = cd$in_tissue,
#> rd10xV+ sample_id = cd$sample_id)
#> sample_id
#> in_tissue section1 section2
#> FALSE 28 27
#> TRUE 22 22
#>
#> rd10xV> # view available images
#> rd10xV> imgData(spe)
#> DataFrame with 2 rows and 4 columns
#> sample_id image_id data scaleFactor
#> <character> <character> <list> <numeric>
#> 1 section1 lowres #### 0.0510334
#> 2 section2 lowres #### 0.0510334
# 'SpatialImage' accession
(spi <- getImg(spe))
#> 576 x 600 (width x height) StoredSpatialImage
#> imgSource():
#> /tmp/RtmppdZi9A/temp_libpathcf263c794d1/SpatialExperiment/extdata/10xVisium/sect
#> ion1/outs/spatial/tissue_lowres_image.png
plot(imgRaster(spi))
 # remove an image
imgData(spe)
#> DataFrame with 2 rows and 4 columns
#> sample_id image_id data scaleFactor
#> <character> <character> <list> <numeric>
#> 1 section1 lowres #### 0.0510334
#> 2 section2 lowres #### 0.0510334
spe <- rmvImg(spe,
sample_id = "section1",
image_id = "lowres")
imgData(spe)
#> DataFrame with 1 row and 4 columns
#> sample_id image_id data scaleFactor
#> <character> <character> <list> <numeric>
#> 1 section2 lowres #### 0.0510334
# add an image
url <- "https://i.redd.it/3pw5uah7xo041.jpg"
spe <- addImg(spe,
sample_id = "section1",
image_id = "pomeranian",
imageSource = url,
scaleFactor = NA_real_,
load = FALSE)
# extract image
img <- imgRaster(spe,
sample_id = "section1",
image_id = "pomeranian")
plot(img)
# remove an image
imgData(spe)
#> DataFrame with 2 rows and 4 columns
#> sample_id image_id data scaleFactor
#> <character> <character> <list> <numeric>
#> 1 section1 lowres #### 0.0510334
#> 2 section2 lowres #### 0.0510334
spe <- rmvImg(spe,
sample_id = "section1",
image_id = "lowres")
imgData(spe)
#> DataFrame with 1 row and 4 columns
#> sample_id image_id data scaleFactor
#> <character> <character> <list> <numeric>
#> 1 section2 lowres #### 0.0510334
# add an image
url <- "https://i.redd.it/3pw5uah7xo041.jpg"
spe <- addImg(spe,
sample_id = "section1",
image_id = "pomeranian",
imageSource = url,
scaleFactor = NA_real_,
load = FALSE)
# extract image
img <- imgRaster(spe,
sample_id = "section1",
image_id = "pomeranian")
plot(img)
 ###################
# transformations #
###################
# clockwise rotation
spe1 <- rotateImg(spe,
degrees = 90) # first image
spe2 <- rotateImg(spe,
sample_id = TRUE,
image_id = TRUE,
degrees = 90) # all images
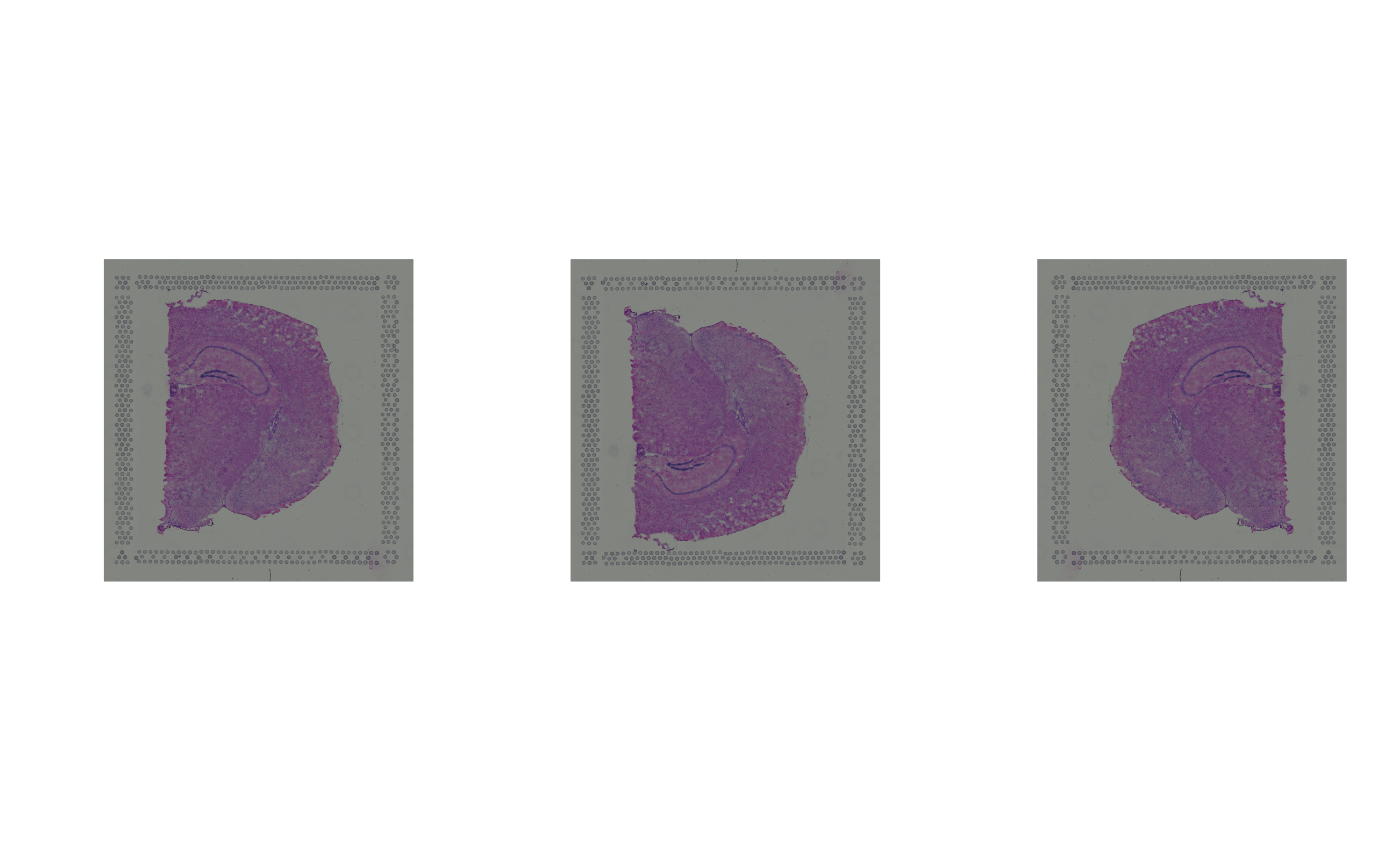
par(mfrow = c(1, 3))
plot(imgRaster(spe))
plot(imgRaster(spe1))
plot(imgRaster(spe2))
###################
# transformations #
###################
# clockwise rotation
spe1 <- rotateImg(spe,
degrees = 90) # first image
spe2 <- rotateImg(spe,
sample_id = TRUE,
image_id = TRUE,
degrees = 90) # all images
par(mfrow = c(1, 3))
plot(imgRaster(spe))
plot(imgRaster(spe1))
plot(imgRaster(spe2))
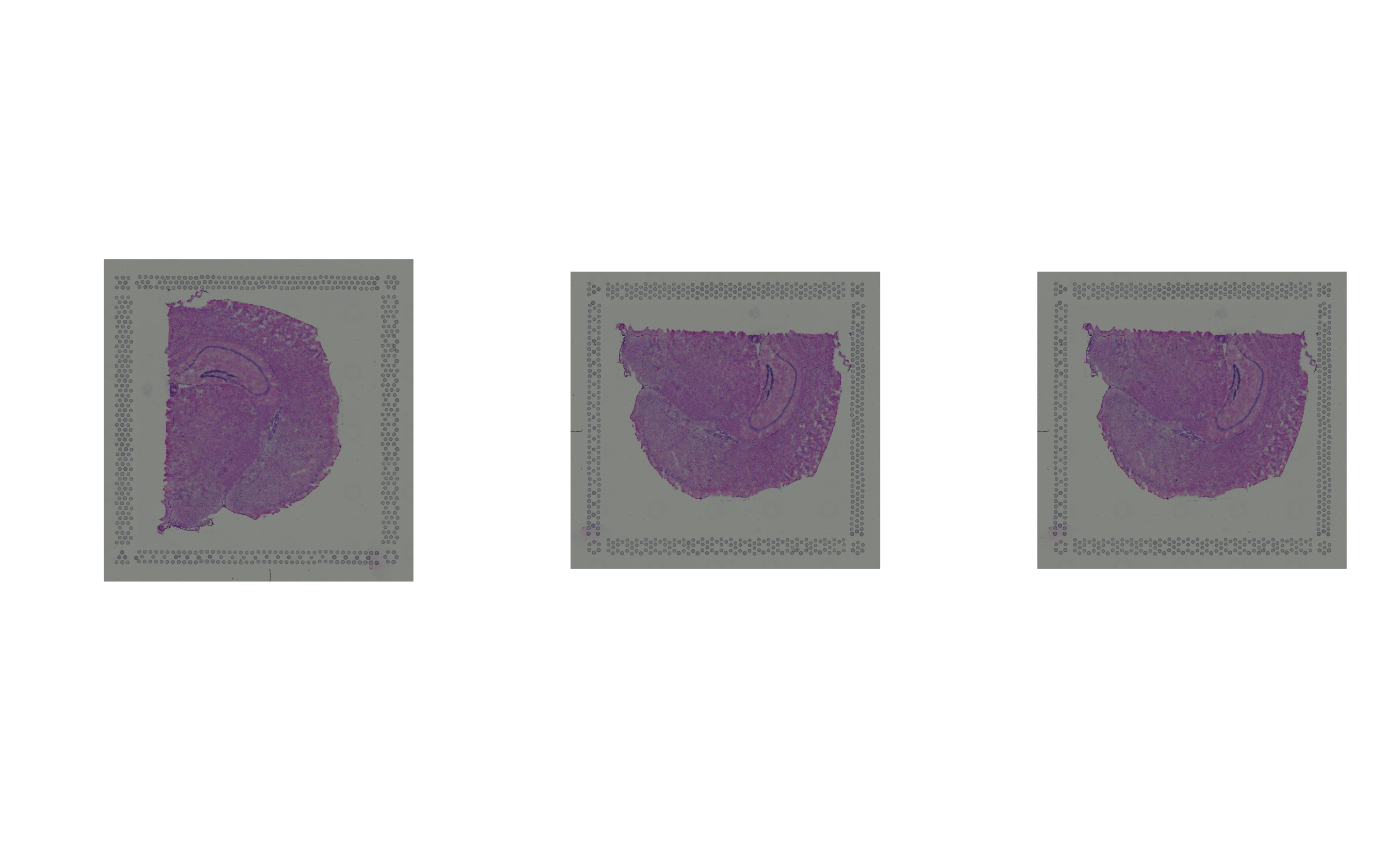 # horizontal/vertical mirroring
spe1 <- mirrorImg(spe, axis = "h")
spe2 <- mirrorImg(spe, axis = "v")
par(mfrow = c(1, 3))
plot(imgRaster(spe))
plot(imgRaster(spe1))
plot(imgRaster(spe2))
# horizontal/vertical mirroring
spe1 <- mirrorImg(spe, axis = "h")
spe2 <- mirrorImg(spe, axis = "v")
par(mfrow = c(1, 3))
plot(imgRaster(spe))
plot(imgRaster(spe1))
plot(imgRaster(spe2))