The SpatialImage class hierarchy provides representations of images
from a variety of sources. It is used by the SpatialExperiment
class to manage the loading of images across multiple studies.
Constructor
SpatialImage(x, is.url) will return a SpatialImage object.
The class of the object depends on the type of x:
If
xis a raster object, aLoadedSpatialImageis returned. This represents an image that is fully realized into memory, where the raster representation is stored inside the output object.If
xis a string andis.url=TRUEor it starts with"http://","http://"or"ftp://", aRemoteSpatialImageis returned. This represents an image that is remotely hosted and retrieved only on request.If
xis a string andis.url=TRUEor it does not start with a URL-like prefix, aStoredSpatialImageis returned. This represents an image that is stored in a local file and is loaded into memory only on request.
Getting the raster image
For a SpatialImage object x, imgRaster(x, ...)
will return a raster object (see ?as.raster).
This is effectively a matrix of RGB colors for each pixel in the image.
For a StoredSpatialImage object x, additional arguments
in ... are passed to image_read.
This controls how the image is read into memory.
For a RemoteSpatialImage object x, the image file is first
downloaded before the raster is returned. Here, ... may contain an
extra cache argument, which should be a BiocFileCache object
(from the BiocFileCache package) specifying the file cache location.
The default location is determined by
options("SpatialExperiment.remote.cache.path"),
otherwise it defaults to a subdirectory in the R temporary directory.
Any further named arguments in ... are passed to image_read.
as.raster(x, ...) is the same as imgRaster(x, ...).
In-memory caching
For StoredSpatialImage and RemoteSpatialImage objects,
loading the image with imgRaster will automatically
store the loaded raster object in an in-memory cache.
Any subsequent imgRaster call will retrieve the raster
from the cache, avoiding costly retrieval from the file system.
The cache policy is to evict the least recently used images when a new image would be added that exceeds the maximum cache size. If the new image by itself exceeds the maximum cache size, all images are evicted from the cache to trigger garbage collection and free up memory.
By default, the maximum size of the cache is 4 GB. This can be
modified by setting options("SpatialExperiment.cache.size")
to some number of bytes, e.g., 2^32.
Transformations
Two basic image transformations are currently
supported for any SpatialImage x, namely,
rotateImg(x, degrees) for clockwise (degrees > 0) and
counter-clockwise (degrees < 0) rotation, and
mirrorImg(x, axis) for horizontal (axis = "h") and
vertical (axis = "v") mirroring.
Note that, both rotateImg() and mirrorImg() operate
on the raster matrix of the input SpatialImage.
Thus, any SpatialImage will automatically be coerced
into a LoadedSpatialImage upon rotation/mirroring.
Other methods
dim(x) will return an integer vector of length 2,
containing the width and height of the image in pixels.
Note that this calls imgRaster under the hood and thus
may interact with the file and memory caches as described above.
For any SpatialImage x, as(x, "LoadedSpatialImage") will
create a LoadedSpatialImage containing an in-memory raster object.
For a RemoteSpatialImage x, as(x, "StoredSpatialImage") will
create a StoredSpatialImage pointing to the file cache location.
Examples
path <- system.file(
"extdata", "10xVisium", "section1", "outs", "spatial",
"tissue_lowres_image.png", package="SpatialExperiment")
spi <- SpatialImage(path)
plot(imgRaster(spi))
# the following operations all use the cache
# so there is no need to reload the image
nrow(spi)
#> [1] 600
ncol(spi)
#> [1] 576
plot(as.raster(spi))
# coercing to an explicitly in-memory raster
spi <- as(spi, "LoadedSpatialImage")
plot(as.raster(spi))
###################
# transformations #
###################
# (counter-)clockwise rotation
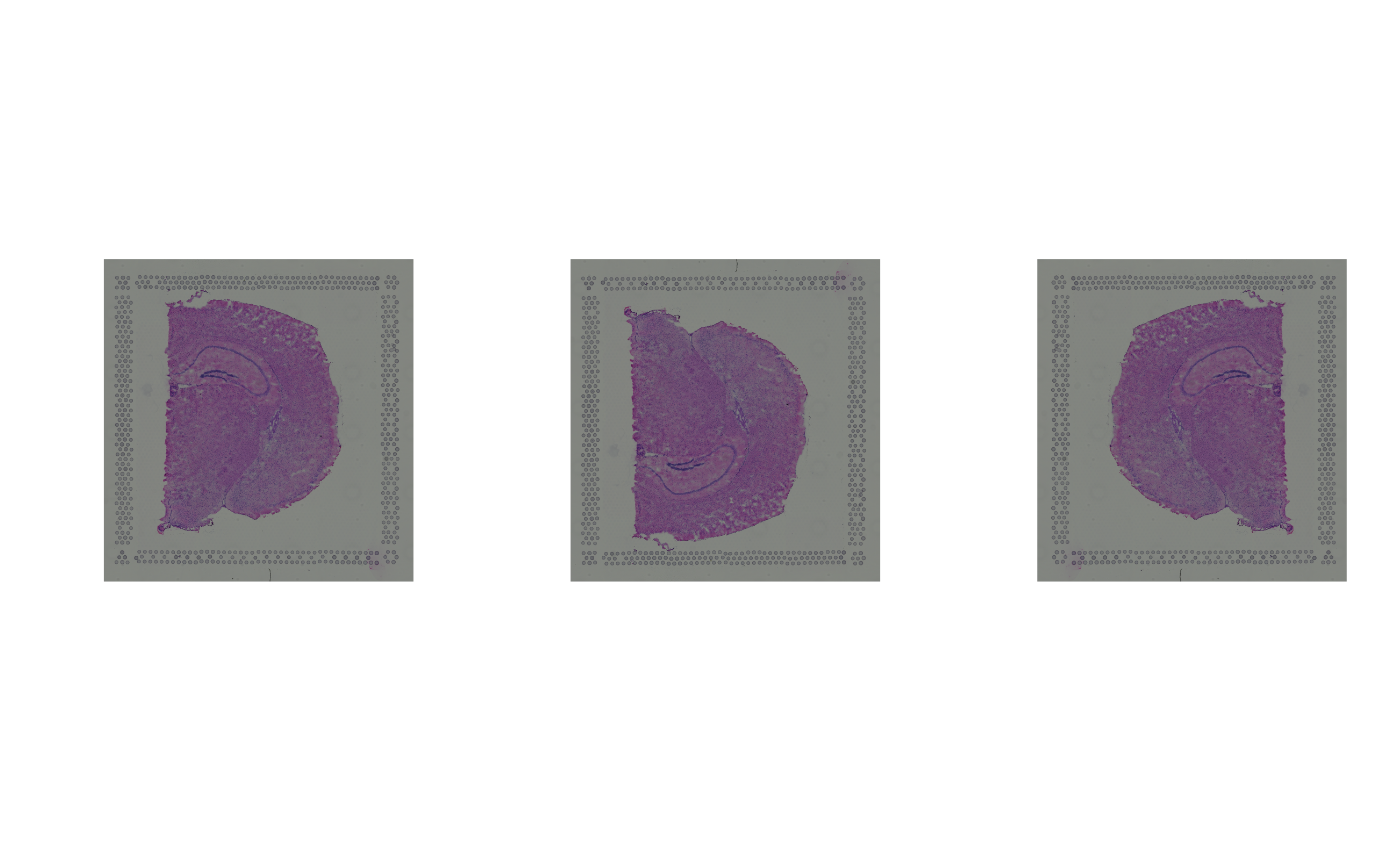
spi1 <- rotateImg(spi, degrees = +90)
spi2 <- rotateImg(spi, degrees = -90)
par(mfrow = c(1, 3))
plot(as.raster(spi))
plot(as.raster(spi1))
plot(as.raster(spi2))
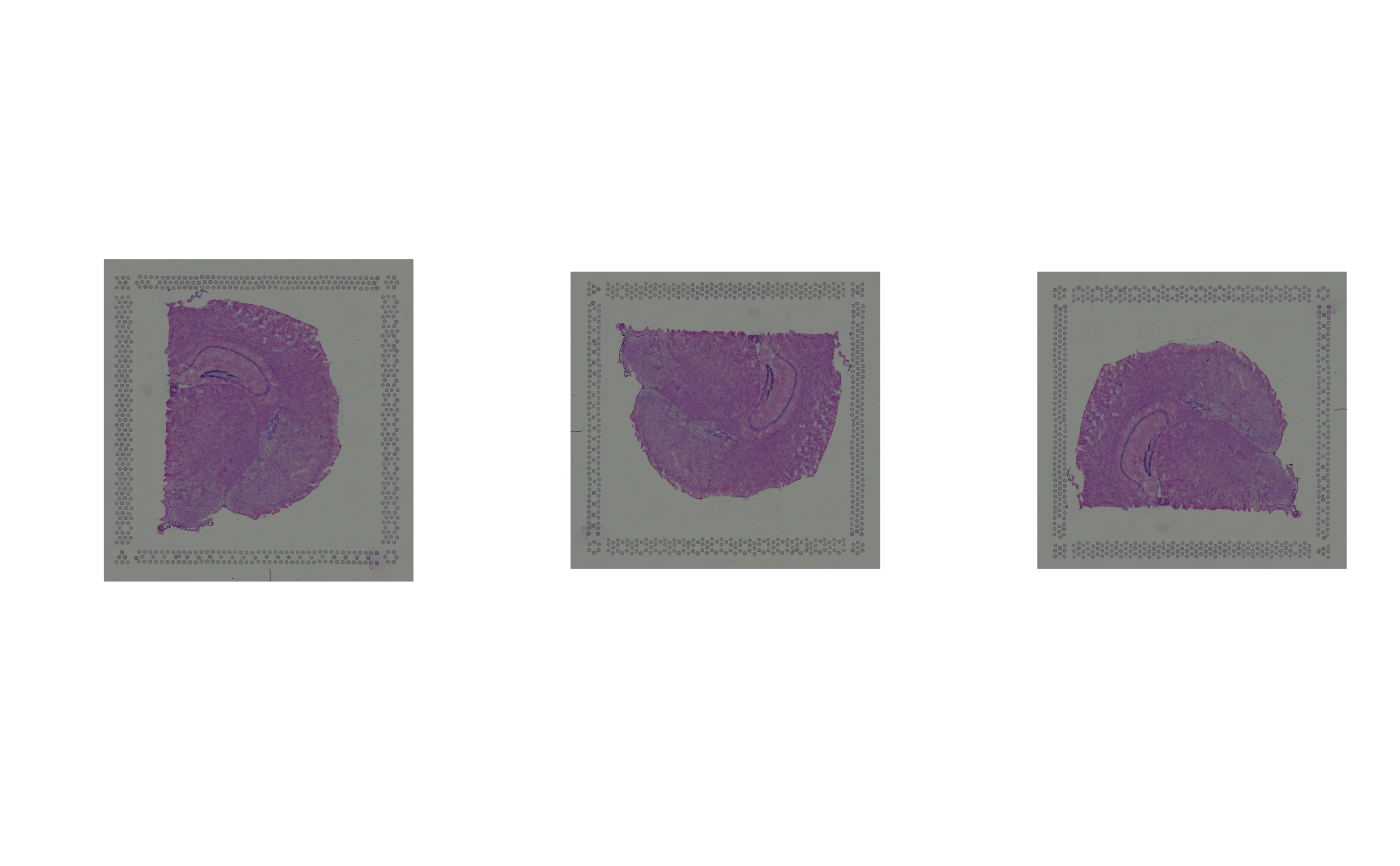 # horizontal/vertical mirroring
spi1 <- mirrorImg(spi, axis = "h")
spi2 <- mirrorImg(spi, axis = "v")
par(mfrow = c(1, 3))
plot(as.raster(spi))
plot(as.raster(spi1))
plot(as.raster(spi2))
# horizontal/vertical mirroring
spi1 <- mirrorImg(spi, axis = "h")
spi2 <- mirrorImg(spi, axis = "v")
par(mfrow = c(1, 3))
plot(as.raster(spi))
plot(as.raster(spi1))
plot(as.raster(spi2))